

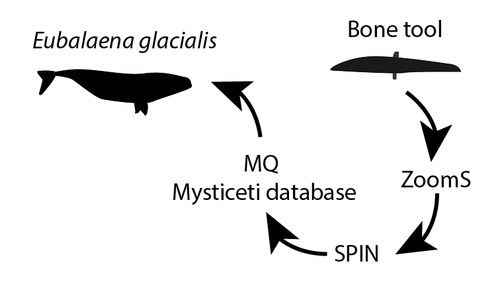
Proteomics is an increasingly applied field of study in archaeology. The characterisation of proteins in ancient biomaterials has been used extensively to determine the sex of certain animals (from dental enamel) or to identify species from non-diagnostic bone pieces or fragments of organic materials (glues and residues, for example). Paleoproteomics has been accompanied by methodological developments, in particular to reduce the size of samples affected by destructive analyses and to refine the level of species determination. The article by Joannes Dekker and colleagues (2024) - "Palaeoproteomic identification of a whale bone tool from Bronze Age Heiloo, the Netherlands" - provides a relevant and innovative example, incorporating ZooMS and SPIN techniques as well as the creation of a database of new reference collagens (cetaceans) specific to the site's natural environment (North Sea coast). The interest of this study also lies in the contribution of a use-wear analysis carried out prior to the sampling. This comparison of multidisciplinary data is essential for understanding the links between man and his natural environment and the technical and economic production that is closely linked to it. The tool studied (ca. 1500 BCE) comes from a coastal Bronze Age site in the Netherlands, where the economy was highly diversified, involving the exploitation of wild and domestic animals in both terrestrial and marine environments. The study shows that the bone of a North Atlantic right whale (Eubalaena glacialis) was shaped into a tool that was probably used to process plant fibres. This discovery supports other studies highlighting the intensive and non-opportunistic exploitation of whales in the North Sea since the Pleistocene.
Dekker, J. A. A., Mylopotamitaki, D., Verbaas, A., Sinet-Mathiot, V., Presslee, S., McCarthy, M. L., Olsen, M. T., Olsen, J. V., van den Hurk, Y., Brattinga, J. & Welker, F. (2024) Palaeoproteomic identification of a whale bone tool from Bronze Age Heiloo, the Netherlands. BioRxiv, ver. 2 peer-reviewed and recommended by Peer Community in Archaeology. https://doi.org/10.1101/2024.04.15.589626
Modifications have been taken into account.
The article is clear and accurate.
The authors have modified the manuscript and responded clearly to all requests. This version of the manuscript is fine for me and ready for publication.
DOI or URL of the preprint: https://doi.org/10.1101/2024.04.15.589626
Version of the preprint: 1
Dear colleagues
Your manuscript has been examined by two reviewers; the decision is minor revision
Please find below the reviewers’ comments and suggestions. Please consider carefully these remarks, respond to all comments individually and provide a new version of the manuscript with the track changes visible.
Reviewer 1 request particularly more information about the archaeological context that led to the choice of this particular sample
Reviewer 2 requests particularly more information about the state of the art
Both reviewers stress on need of further explanations on the sampling, the method as well as analytical information
We propose to receive your revised version by the 5th of July 2024. If you need an extra delay please let us know.
Yours sincerely
· Title and abstract
Does the title clearly reflect the content of the article? Yes
Does the abstract present the main findings of the study? Yes,
· Introduction
Are the research questions/hypotheses/predictions clearly presented? Yes
Does the introduction build on relevant research in the field? Yes
· Materials and methods
Are the methods and analyses sufficiently detailed to allow replication by other researchers? [ ] No. explanation below
Are the methods and statistical analyses appropriate and well described? Yes
· Results
In the case of negative results, is there a statistical power analysis (or an adequate Bayesian analysis or equivalence testing)? Yes
Are the results described and interpreted correctly? Yes,
· Discussion
Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? Yes
Are the conclusions adequately supported by the results (without overstating the implications of the findings)? Yes
The article is well written. Some points could be improved for greater clarity.
The discussion on the importance of the proteomics database is clear.
The repository of ZooMS, SPIN and script data is a good thing for the community.
Introduction.
Sentence : “Ancient DNA analyiss is understood……” you can indicate that the quantity of material required for analysis is too great for these archaeological objects.
In the next sentence, for paleproteomics, you could indicate that there are minimally invasive methods available.
For SPIN analysis, which is a shotgun analysis, it is possible to obtain phylogeny information.
It would be interesting to add a paragraph on the use of proteomics on archaeological tools. There are examples in the literature.
Materials & methods
Page 3, Figure 1 c. can you indicate the position of the bone
Page 3, Why 2 ZooMS analyses? Why take 3 samples of 38 mg? Current methods show that 5-1 mg is sufficient. please clarify these points
Page 4, ZooMS
NH4HCO3 ----- > incorrect number format, NH4HCO3
Page 5, 50µL ----- > 50 µL a space is missing
Page 5, Missing MALDI information: mass range, positive mode, calibration....
Page 5, a space is missing “50 mM”
Page 5, SPIN. did you measure the quantity of peptides injected? please indicate
Page 5, Maxquant:
Custom database of COL 1 form NCBI? Or Swissprot? These are the complete or mature sequences (with ou without signal peptide)?
Why you used 2 version of Maxquant?
Why you used specific tryptic cleavage? Using the semi-trypsin mode, the number of peptides identified increases. Did you test it?
Bioinformatics data analysis, Mass spectrometric data. “default settings and protein sequence database” can you explain. Database of which proteins? (collagen? Others?)
Results
ZooMS, page 8. Why didn't you watch the deamidation?
SPIN page 10. You have a database of cetacean collagen and you identify bos. There must be information missing somewhere.
Page 11, “For each of the five SAPs the number of peptide spectrum matches (PSMs)” have you done a blast analysis to check the specificity of the sequence?
Discussion
Page 15 “Three proteomic workflows”. You have used 2 different workflows ZooMS and SPIN.
You don't mention de novo mass spectrometry analysis. Do you think these could help when analyzing samples without protein sequences? not all laboratories can afford genome sequencing.
The title clearly reflect the content of the article and the abstract present the main findings of the study. The research questions are clearly presented and the introduction build on relevant research in the field. The methods and analyses are sufficiently detailed to allow replication by other researchers, appropriate and well described, I only suggest some small clarifications. The results are described and interpreted correctly, I just need some explanations. The authors appropriately emphasized the strengths and limitations of their study and the conclusions are adequately supported by the results.
Download the review